Bridging Dynamic Network Data and Epidemic Simulation: A Computational Framework
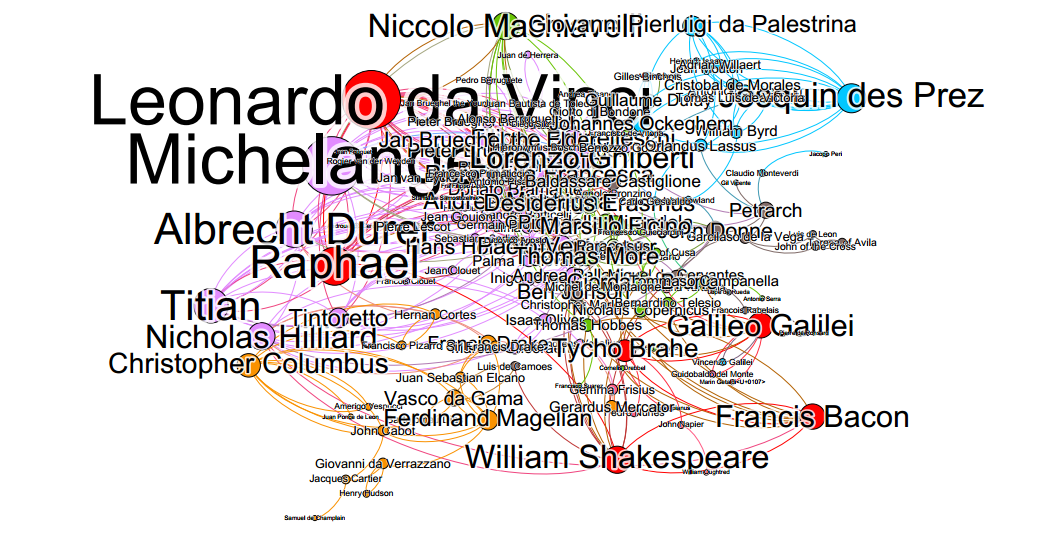
Click hereModeling the Renaissance: A Historical Art Network Using Computational Methods in R and Gephi
Click here The easiest way to install tensorflow by using conda
As I mentioned in the previous post, it is tricky to install tensorflow. The easiest way is using conda.
- Create a virtual environment.
conda create -n tensorflow-env
# If you want remove the environment
conda remove -n tensorflow-env --all - In the environment, install
tensorflow-gpuortensorflowand check it.
conda activate tensorflow-env
conda install tensorflow-gpu
# Check
import tensorflow as tf
tf.__version__ - If you want to use
tensorflowin the Jupyter notebook, install ipython and Jupyter notebook in the environment. After checkingtensorflowin the ipython, then usectrl + dto exit.
conda install ipython
conda install jupyter notebook
# Check in the jupyter notebook When insert Python chunk into R markdown, matplotlib error
Sometimes R Studio cannot find matplotlib or a Python library when we try to insert Python chunk into R markdown.
Everything is correct and the code works well in the Jupyter. But it does not work in the R markdown.
We need to use a different way to install the Python library
SOLUTION :
In R chunk, py_install(packages = "matplotlib")
In Python chunk, import os
os.environ['QT_QPA_PLATFORM_PLUGIN_PATH'] = '(anaconda or miniconda path) ...conda3/Library/plugins/platforms'
Fail to create Virtual Environment for Python in the Window Powershell
To create your Virtual Environment,
Type : conda create -n env python = 3.8 and conda activate env
Then, you should find (env) in the Window Powershell
If you cannot find it, you failed to create your Python Vitural Environment. Interestingly, it can be available only in cmd.
I think Window Powershell has a compatibility issue with Python.
SOLUTION:
In your Powershell, Set-ExecutionPolicy -Scope "CurrentUser" -ExecutionPolicy "Unrestricted"
R Markdown issue about TinyTex and MikTex : Fail to compile
Install tinytex package in your R. Then tinytex::install_tinytex() Now, you may face a compiling error. The easiest way to solve this is to uninstall MikTex. Quit and reopen the R session. Then tinytex::install_tinytex() again. If you succeed in compiling your tex file. Now reinstall MikTex. When you use a new tex package in R markdown, it also can make another compiling error. If R markdown creates a tex file, Open the tex file in your LaTex editor. Then your MikTex automatically updates the new package.
R package install Error from github : devtools error
When you install a new package from github, a devtools error will be a challenge.SOLUTION: Install the recent version Rtools. Next step is changing .Renviron file. To do this, in R or Rstudio, put the following line : writeLines('PATH="${RTOOLS40_HOME}\\usr\\bin;${PATH}"', con = "~/.Renviron") Then Restart R, and verify the PATH in .Renviron with Sys.which("make")
Easy ways to open ipynb files in Jupyter Notebook
Go to the folder containging ipynb files. On the address cell of the File explorer(Windows), type jupyter notebook (e.g. C:\aaa -> jupyter notebook).If it does not work, you need to change your 'environment variables' of your Windows. Choose change my environment variables in the 'User Account' of the 'Control Panel'. Choose 'Path' in the 'User variables' and click 'Edit'. Add two new paths for python.exe and conda.exe (e.g. C:\User\Anaconda3, C:\User\Anaconda3\Scripts ). Help : Use Anaconda Prompt. where python and where conda
If it does not work, we can change the starting folder. By using the right click, choose "Properties" of Jupyter Notebook's shortcut tab. In the "Target", change "%USERPROFILE%/" to your specific folder. e.g. ...\jupyter-notebook-script.py "%USERPROFILE%/" → ...\jupyter-notebook-script.py "D:/python folder/"
--notebook-dir= jupyter notebook --notebook-dir=./(your working folder)
# For my case
jupyter notebook --notebook-dir=./OneDrive Python : Install Keras and Tensorflow
Keras is a Python library for deep learning modeling. If you are using Python 3.8 and Anaconda, you will face some troubles with installing Keras, because Tensorflow is required to install Keras. It is tricky to install Tensorflow into Python 3.8 due to versions of packages and Python.
Solution : Create a virtual environment for installing Tensorflow and Python 3.8
In the Prompt or Powershell, create a virtual environment with conda :
conda create -n tensorflowenv python 3.7.
After activating the Tensorflow environment,
conda install tensorflow-gpu=2.1.0 -c anaconda
Now, Tensorflow is installed along with Keras. After checking Keras, leave from the virtual environment for Python 3.7 by :
conda deactivate
Help : The virtual environment settings depend on the version of Tensorflow you want to install. [cf] : '-n' means name.
Web scraping with Python : urllib.request.urlopen - HTTP error and solution
When you use urllib.request.uropen to scrape a webpage, you may get some HTTP errors. I suppose the server think you could be a bot and block your scraping.Solution : We need to reset the browser user agent to avoid the blocking. import urllib.request url = 'http://..........' req = urllib.request.Request(url, headers={'User-Agent': 'Mozilla/5.0'}) page = urllib.request.urlopen(req).read() Now, you can scrape the webpage without error messages.
Sweave in R and Latex
Sweave is a good tool for R and Latex users. But you may find an error message when you make your first tex file using Sweave. "Sweave.sty not found." It means no package for Sweave.sty in MikTex. You can find this file in the R folder. C\...\R\share\texmf\texThe easiest way to use this file is as follows: First of all, find and clink 'mo' file in C\...\MiKTex2.9\miktex\bin Second, choose Roots tap. Third, click add and choose C\....\R\share\texmf If you are looking for another option using R and Latex, I recommend R package knitr. It is convenient and you can make a good-looking document
Gephi : Solution to Java error
When you install or upgrade Gephi, your Gephi may not open and show an error message
"Gephi cannot find Java 1.8 or higher"
To solve this issue, first of all find the location of Java in your computer.
For example, C:\Program Files (x86)\Java\jre1.8.0_144
Second, you need to find a file "gephi.conf" in the etc folder of Gephi
Third, open the file with any notepad and emove "#" before jdkhome=.
Then, put the location of Java after jdkhome=.
For example, jdkhome="C:/Program Files (x86)/Java/jre1.8.0_144"
Save the edited file and Run Gephi
Visualizing Airline routes Network
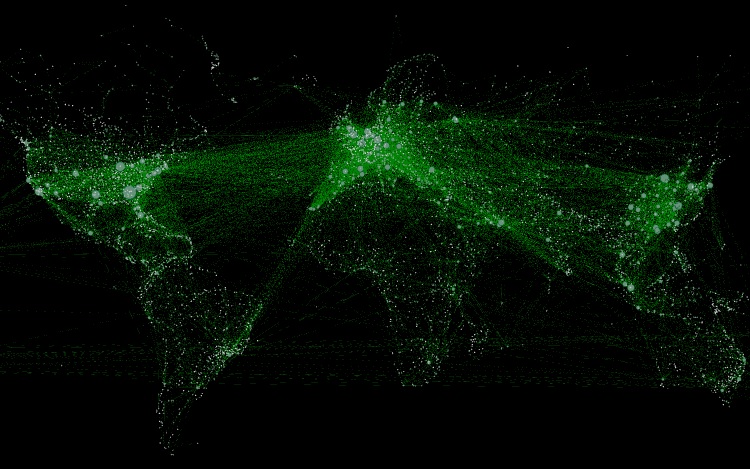 We can get Aitline routes and Airports data from https://openflights.org.
By using Gephi, we can easily visualize the Airline routes data over a map.
In my Youtube channel, you can find my Gephi tutorial video to visualize this Airline routes network.
Tutorial is Here
You can download nodes data and
links data.
We can get Aitline routes and Airports data from https://openflights.org.
By using Gephi, we can easily visualize the Airline routes data over a map.
In my Youtube channel, you can find my Gephi tutorial video to visualize this Airline routes network.
Tutorial is Here
You can download nodes data and
links data.
Four Tutorials on Dynamic(Temporal) Network using R (+ EPI model) and Gephi
I made four tutorials to deal with dynamic network data and visualizing them. The first tutorial is here. I got this data from Vanhems et al., 2013, Estimating Potential Infection Transmission Routes in Hospital Wards Using Wearable Proximity Sensors. You can download the dataset I used in the tutorial : nodes data and edges data.
In the second tutorial, I teach how to use a gexf file in Gephi. You can find Tutorial part 2 (Import with static files) here. This gexf data comes from Isella et al., 2011, What's in a crowd? Analysis of face-to-face behavioral networks. You can download the data of the tutorial : gexf files
The third tutorial uses the dataset of the first one. I analyze the dynamic network data with R. The tutorial is here . You can check the R code in my gist
The fourth tutorial continues from the first and the third. I assume a virus is spreading in a hospital. I simulate this contagious event with EPImodel(R package) and Vanhems et al.'s dataset. The tutorial is here. You can download the R dataset : R data . The R code is in my gist
Understanding Netlogo Code
HELP: From Netlogo 6.0 or later, "?" is not used anymore, and the R-extension bugs are hardly found.
reduce
'reduce' is a little complicated.
For example, show reduce [?1] [1 2 3] => 1 , show reduce [?2] [1 2 3] => 3
Now, what value can we get from show reduce [?2] [1 2 3 4 5] ?
It will be 5.
'reduce' computes the given task from left to right of the list.
In the question, [?2] means finds the second value in the list.
For the first round, 'reduce' considers only [1] of the list. Thus, the first value in the list is 1 and the second value is also 1.
In the Next round, 'reduce' can consider one more value in the list,[1,2]. The second value is now 2.
Then, the list 'reduce' considers will be [2,3]. We can realize that 'reduce' looks at only 2 values. In [2,3], the second value is 3.
The next list is [3,4], and the second value is 4.
Through this process, we can get 5 in the final round.
When can we use 'reduce'? This is useful for coding math, like Σ Xi.
When X = [1,2,3,4,5],
Σ Xi can be rewritten as reduce + [1 2 3 4 5].
Max Xi can be expressed by reduce [ifelse-value (?1 > ?2) [?1] [?2]] [1 2 3 4 5] .
R-extension error and Solution
r-extension
r-extension is a very useful tool to connect Netlogo to R. We can use R in the Netlogo model with this extension. But, it is a little bit tricky to use it. I had some trouble with loading a package in the Netlogo. Netlogo could not load R package. Here are two solutions
Use R function : Create a R function for your purpose. For example, my function is `mykurtosis'.
In the Netlogo, Type : let mykurtosis (word "source('C:/..(path)../mykurtosis.R')")
r:eval mykurtosis
Now, you can use your R function in your Netlogo model
Change package Path :
Type r:interactiveShell into NetLogo's Command Center.
In the interactiveShell, Type .libPaths()
Check the path for R package. If it is different from yours, do as follows :
In the Netlogo code, Type :
r:put "path" "C:/...(your R library path) .../Documents/R/R-3.2/library"
r:eval ".libPaths(c(path,.libPaths()))"
Now, you can load your R package in the Netlogo.
RNetlogo error and Solution
RNetlogo
Another extension for R and Netlogo is RNetlogo. With this extension, we can use Netlogo in R interface.
But you may face some error messages. One of them is as follows :
WARNING: Could not open/create prefs root node Software\JavaSoft\Prefs at root 0x80000002....
SOLUTION:
First, go to Start menu and type regedit
Login as the administrator, and find HKEY_LOCAL_MACHINE\Software\JavaSoft
Click right button and create key, Prefs.
We need HKEY_LOCAL_MACHINE\Software\JavaSoft\Prefs .