PyNetlogo Tutorial
PyNetlogo is a python package to link Netlog to Python. The latest version is 0.3. I do not think it is well managed after the release of version 0.3. There are three important webpages as follows:
Install and Virtual Environment
I strongly recommend creating a virtual environment for a new package. JPype package is more stable in Pyhton 3.8 or older.
conda create -n netlogo-env python=3.8
# For JPype
conda install -c conda-forge jpype1
# For PyNetLogo
pip install pynetlogo Import Libraries
import pandas as pd
import numpy as np
from scipy import stats
import matplotlib.pyplot as plt
import seaborn as sns
import pyNetLogo
sns.set_style('white')
sns.set_context('talk')
%matplotlib inline
Get Data from Netlogo simulation: .command and .report
# open Netlogo
netlogo = pyNetLogo.NetLogoLink(gui=True)
# Load the Model
netlogo.load_model('C:/Program Files/NetLogo 6.1.1/.../Wolf Sheep Predation.nlogo')
# Setup and Choose model-version = "sheep-wolves-grass"
netlogo.command('setup')
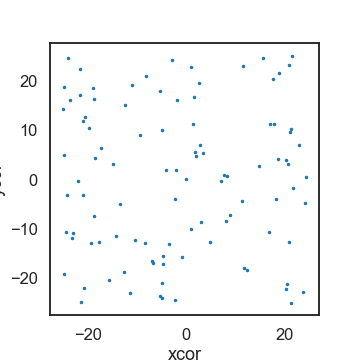
# Get the coordinates of agents(sheep) from Netlogo
x = netlogo.report('map [s -> [xcor] of s] sort sheep')
y = netlogo.report('map [s -> [ycor] of s] sort sheep')
# Make a plot
fig, ax = plt.subplots(1)
ax.scatter(x, y, s=4)
ax.set_xlabel('xcor')
ax.set_ylabel('ycor')
ax.set_aspect('equal')
fig.set_size_inches(5,5)
plt.show()
Repeat the model simulation: Using .command or .repeat_report
# Run 100ticks
netlogo.command('repeat 100 [go]')
# or
# Run 100 ticks and Report 'count wolves'
netlogo.repeat_report(['count wolves'], 100, go='go')
Import data to the Netlogo model : .write_NetLogo_attriblist
# ex_data is a pandas DataFrame having three columns : col1, col2, and col3.
ex_data[['col1','col2','col3']]
# Import the data to col1, col2 and col3 attributes of the 'breed'
netlogo.write_NetLogo_attriblist(ex_data[['col1','col2','col3']], 'breed')
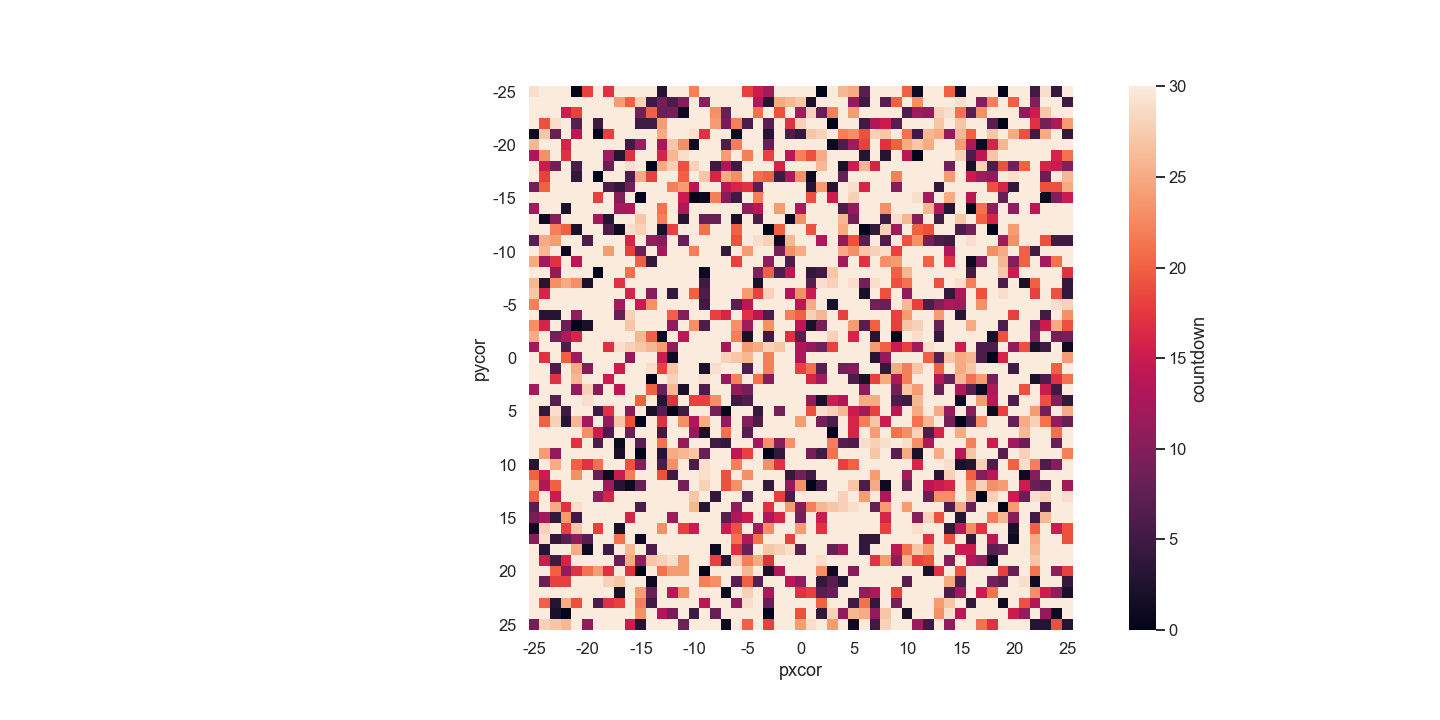
.patch_report is not Working.
To get data from patches, we need to use .patch_report.
But it gives an error message. Alternatively, I use .report.
# Error
countdown_df = netlogo.patch_report('countdown')
# Alternative way
countdown_df=netlogo.report('map [s -> [countdown] of s] sort patches')
# Reshape for 51 by 51 square
countdown_df=countdown_df.reshape(51,51)
# Convert to DataFrame
countdown_df = pd.DataFrame(countdown_df)
# Change index and column names
countdown_df.columns=[list(range(-25,26))]
countdown_df.index=[list(range(-25,26))]
# Make a plot
fig, ax = plt.subplots(1)
patches = sns.heatmap(countdown_df, xticklabels=5, yticklabels=5,
cbar_kws={'label':'countdown'}, ax=ax)
ax.set_xlabel('pxcor')
ax.set_ylabel('pycor')
ax.set_aspect('equal')
fig.set_size_inches(20,10)
plt.show()
Export patch data and change patch data : .to_excel and .patch_set>
countdown_df.to_excel('countdown.xlsx')
netlogo.patch_set('countdown', countdown_df.max()-countdown_df)
Sensitivity Analysis - using SALib
I think the PyNetlogo website needs to be updated about the Sensitivity analysis part because the codes are not working. The SALib has two views for the analysis. One is calledDirect view, and the other is Load balanced view.
I cannot find an alternative way to use the latter which the website used.
In this tutorial, I use SALib Direct view method for a sensitivity analysis.
Import two functions for sampling and analysis.
from SALib.sample import saltelli
from SALib.analyze import sobol
- Determine parameters and their range
problem = {
'num_vars': 6,
'names': ['random-seed',
'grass-regrowth-time',
'sheep-gain-from-food',
'wolf-gain-from-food',
'sheep-reproduce',
'wolf-reproduce'],
'bounds': [[1, 100000],
[20., 40.],
[2., 8.],
[16., 32.],
[2., 8.],
[2., 8.]]
}
- Sampling parameter sets using
saltellisample function
n = 1000
param_values = saltelli.sample(problem, n, calc_second_order=True)
# We can get 14,000 parameter sets.
- Run the
Target modelwith the parameter sets
# Definition for simulation
def simulation(experiment):
#Set the input parameters
for i, name in enumerate(problem['names']):
if name == 'random-seed':
#The NetLogo random seed requires a different syntax
netlogo.command('random-seed {}'.format(experiment[i]))
else:
#Otherwise, assume the input parameters are global variables
netlogo.command('set {0} {1}'.format(name, experiment[i]))
netlogo.command('setup')
#Run for 100 ticks and return the number of sheep and wolf agents at each time step
counts = netlogo.repeat_report(['count sheep','count wolves'], 100)
results = pd.Series([counts['count sheep'].values.mean(),
counts['count wolves'].values.mean()],
index=['Avg. sheep', 'Avg. wolves'])
return results
# DO NOT RUN this in the Jupyter Notebook
# Run it in the Terminal
# I choose 8,but you can choose 4
ipcluster start -n 8
import ipyparallel as ipp
client = ipp.Client() # client - setup
client.ids
# Allow direct view to use all engines
direct_view = client[:]
# For Parallel Engine access
import os
direct_view.push(dict(cwd=os.getcwd()))
direct_view.push(dict(problem=problem))
# Run Engines
%%px
# Load the Netlogo model
import os
os.chdir(cwd)
import pyNetLogo
import pandas as pd
netlogo = pyNetLogo.NetLogoLink(gui=True)
netlogo.load_model('Wolf Sheep Predation_pynetlogo.nlogo')
# Run the Model with parallel Engines
results = pd.DataFrame(direct_view.map_sync(simulation, param_values))
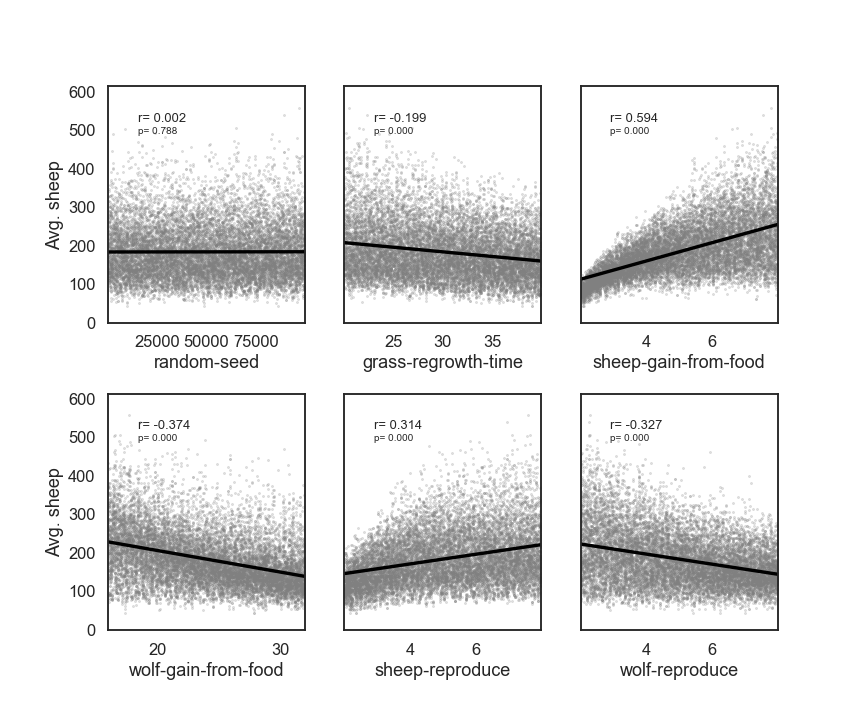
# Make Plots for the results
import scipy
fig, ax = plt.subplots(2, 3, sharey=True)
y = results['Avg. sheep']
for i, a in enumerate(ax.flatten()):
x = param_values[:,i]
sns.regplot(x=x, y=y, ax=a, ci=None, color='k',scatter_kws={'alpha':0.2, 's':4, 'color':'gray'})
pearson = scipy.stats.pearsonr(x, y)
a.annotate("r= {:.3f}".format(pearson[0]), xy=(0.15, 0.85), xycoords='axes fraction',fontsize=13)
a.annotate("p= {:.3f}".format(pearson[1]), xy=(0.15, 0.8), xycoords='axes fraction',fontsize=10)
if divmod(i,3)[1]>0:
a.get_yaxis().set_visible(False)
a.set_xlabel(problem['names'][i])
a.set_ylim([0, 1.1*np.max(y)])
fig.set_size_inches(12,10,forward=True)
fig.subplots_adjust(wspace=0.2, hspace=0.3)
plt.show()
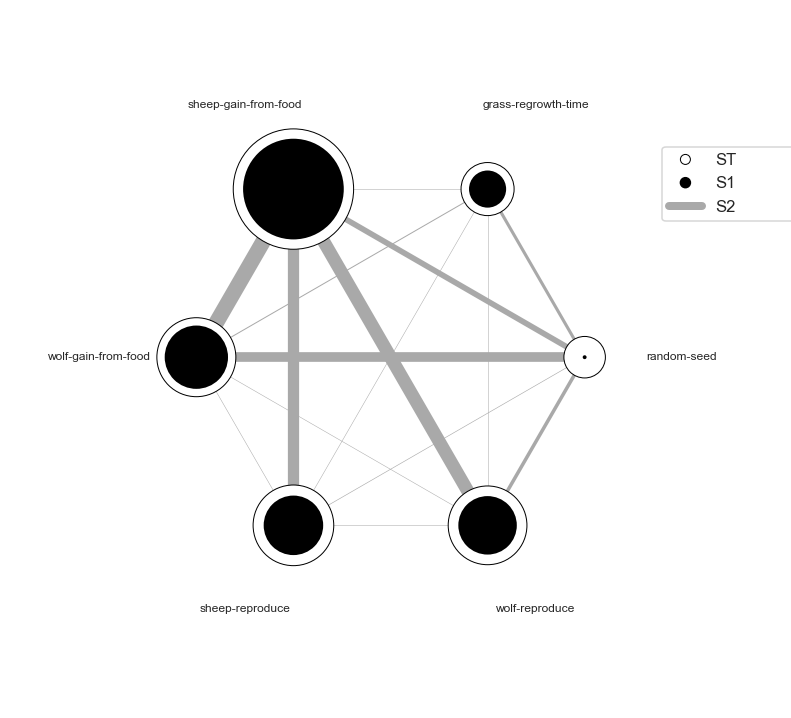
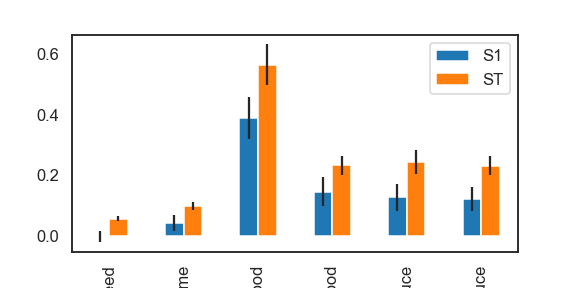
- Sensitivity analysis using
SobolfunctionSobol function generates three indices : S1 : The contribution to the output variance by single parameter. Single parameter → the Output. S2 : The interaction of two parameters → the Output. ST : One or more parameters → the Output.
Si = sobol.analyze(problem, results['Avg. sheep'].values, calc_second_order=True, print_to_console=True)
# For S1 and ST
Si_filter = {k:Si[k] for k in ['ST','ST_conf','S1','S1_conf']}
Si_df = pd.DataFrame(Si_filter, index=problem['names'])
# For S2
Si2_filter = {k:Si[k] for k in ['S2', 'S2_conf']}
Si2_df = pd.DataFrame(Si2_filter['S2'], columns=problem['names'],index=problem['names'])
Si2_conf = pd.DataFrame(Si2_filter['S2_conf'], columns=problem['names'],index=problem['names'])
fig, ax = plt.subplots(1)
indices = Si_df[['S1','ST']]
err = Si_df[['S1_conf','ST_conf']]
indices.plot.bar(yerr=err.values.T, ax=ax)
fig.set_size_inches(8,4)
plt.show()
import itertools
from math import pi
#sobol_indices = Si
def normalize(x, xmin, xmax):
return (x-xmin)/(xmax-xmin)
def plot_circles(ax, locs, names, max_s, stats, smax, smin, fc, ec, lw,
zorder):
s = np.asarray([stats[name] for name in names])
s = 0.01 + max_s * np.sqrt(normalize(s, smin, smax))
fill = True
for loc, name, si in zip(locs, names, s):
if fc=='w':
fill=False
else:
ec='none'
x = np.cos(loc)
y = np.sin(loc)
circle = plt.Circle((x,y), radius=si, ec=ec, fc=fc, transform=ax.transData._b,
zorder=zorder, lw=lw, fill=True)
ax.add_artist(circle)
## Create filtered_names, filtered_locs
def filter(sobol_indices, names, locs, criterion, threshold):
if criterion in ['ST', 'S1', 'S2']:
data = sobol_indices[criterion]
data = np.abs(data)
data = data.flatten() # flatten in case of S2
# TODO:: remove nans
filtered = ([(name, locs[i]) for i, name in enumerate(names) if data[i]>threshold])
filtered_names, filtered_locs = zip(*filtered)
elif criterion in ['ST_conf', 'S1_conf', 'S2_conf']:
raise NotImplementedError
else:
raise ValueError('unknown value for criterion')
return filtered_names, filtered_locs
#######
## Main
########
def plot_sobol_indices(sobol_indices, criterion='ST', threshold=0.01):
'''plot sobol indices on a radial plot
Parameters
----------
sobol_indices : dict
the return from SAlib
criterion : {'ST', 'S1', 'S2', 'ST_conf', 'S1_conf', 'S2_conf'}, optional
threshold : float
only visualize variables with criterion larger than cutoff
'''
max_linewidth_s2 = 15#25*1.8
max_s_radius = 0.3
# prepare data
# use the absolute values of all the indices
#sobol_indices = {key:np.abs(stats) for key, stats in sobol_indices.items()}
# dataframe with ST and S1
sobol_stats = {key:sobol_indices[key] for key in ['ST', 'S1']}
sobol_stats = pd.DataFrame(sobol_stats, index=problem['names'])
smax = sobol_stats.max().max()
smin = sobol_stats.min().min()
# dataframe with s2
s2 = pd.DataFrame(sobol_indices['S2'], index=problem['names'],
columns=problem['names'])
s2[s2<0.0]=0. #Set negative values to 0 (artifact from small sample sizes)
s2max = s2.max().max()
s2min = s2.min().min()
names = problem['names']
n = len(names)
ticklocs = np.linspace(0, 2*pi, n+1)
locs = ticklocs[0:-1]
filtered_names, filtered_locs = filter(sobol_indices, names, locs,
criterion, threshold)
####################
####################
# setup figure
####################
#####################
fig = plt.figure()
ax = fig.add_subplot(111, polar=True)
ax.grid(False)
ax.spines['polar'].set_visible(False)
ax.set_xticks(locs) ## Fixed ##
ax.set_xticklabels(names, fontsize=12)
ax.set_yticklabels([])
ax.set_ylim(top=1.4)
legend(ax)
# plot ST
plot_circles(ax, filtered_locs, filtered_names, max_s_radius,
sobol_stats['ST'], smax, smin, 'w', 'k', 1, 9)
# plot S1
plot_circles(ax, filtered_locs, filtered_names, max_s_radius,
sobol_stats['S1'], smax, smin, 'k', 'k', 1, 10)
# plot S2
for name1, name2 in itertools.combinations(zip(filtered_names, filtered_locs), 2):
name1, loc1 = name1
name2, loc2 = name2
weight = s2.loc[name1, name2]
lw = 0.5+max_linewidth_s2*normalize(weight, s2min, s2max)
ax.plot([loc1, loc2], [1,1], c='darkgray', lw=lw, zorder=1)
return fig
from matplotlib.legend_handler import HandlerPatch
class HandlerCircle(HandlerPatch):
def create_artists(self, legend, orig_handle,
xdescent, ydescent, width, height, fontsize, trans):
center = 0.5 * width - 0.5 * xdescent, 0.5 * height - 0.5 * ydescent
p = plt.Circle(xy=center, radius=orig_handle.radius)
self.update_prop(p, orig_handle, legend)
p.set_transform(trans)
return [p]
def legend(ax):
some_identifiers = [plt.Circle((0,0), radius=5, color='k', fill=False, lw=1),
plt.Circle((0,0), radius=5, color='k', fill=True),
plt.Line2D([0,0.5], [0,0.5], lw=8, color='darkgray')]
ax.legend(some_identifiers, ['ST', 'S1', 'S2'],
loc=(1,0.75), borderaxespad=0.1, mode='expand',
handler_map={plt.Circle: HandlerCircle()})
## plot
sns.set_style('whitegrid')
fig = plot_sobol_indices(Si, criterion='ST', threshold=0.005)
fig.set_size_inches(15,10)
plt.show()